class: center, middle, inverse, title-slide .title[ # Data visualization primer ] .subtitle[ ## Making good choices for data visualization ] .author[ ### Guillaume Falmagne ] .date[ ### <br> Oct. 30th, 2024 ] --- # General recommendations (+ choosing plot type) .pull-left[ - A big part of data visualization is choosing the correct type of plot to represent the data you have - You should consider first the **MESSAGE** you want to convey ] .pull-right[  ] .phantom[d] .negspace100[ - Then consider: - **type** of plot (this lecture) - **aesthetics**, taking into account colorblindness (previous lecture) - simplicity/**ease of reading vs exhaustivity** - every element of the plot is a potential visual to convey your message! - the visual should be a **honest** representation of your data/results - There are some (flexible) general rules depending on the nature of the data -- but there is also some trial and error ] --- class: left, top background-image: url(figures/Doumont.jpg) background-position: center right background-size: 38% # Readings [Jean Luc Doumont, *Trees, Maps, and Theorems*](https://www.goodreads.com/book/show/6477103-trees-maps-and-theorems) [Rougier, *Scientific Visualization: Python + Matplotlib*](https://inria.hal.science/hal-03427242/document) [Tufte, *The Visual Display of Quantitative Information*](https://www.edwardtufte.com/book/the-visual-display-of-quantitative-information/) [Tufte, *Envisioning Information*](https://www.edwardtufte.com/book/envisioning-information/) [Ware, *Information Visualization*](https://www.researchgate.net/publication/224285723_Information_Visualization_Perception_for_Design_Second_Edition) [Wilke, *Fundamentals of Data Visualization*](https://clauswilke.com/dataviz/) --- class: inverse, center, middle # Lines and points --- # Lineplots (R) - Implies **continuity** between data points - Ideal for **time-series** data - Can be combined with **points/markers** to highlight individual observations .pull-left[ ``` r library(tidyverse) # Load dataset from github data <- read.table("https://raw.githubusercontent.com/holtzy/data_to_viz/master/Example_dataset/3_TwoNumOrdered.csv", header=T) data$date <- as.Date(data$date) data |> ggplot(aes(x=date, y=value)) + * geom_line(color="#69b3a2") + ylim(0,22000) + annotate(geom="text", x=as.Date("2017-01-01"), y=20089, label="Bitcoin price reached $20k \nat the end of 2017") + annotate(geom="point", x=as.Date("2017-12-17"), y=20089, size=10, shape=21, fill="transparent") + geom_hline(yintercept=5000, color="orange", linewidth=1) + theme_minimal_grid() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-2-1.png" width="90%" /> ] --- # Lineplots (python) .pull-left[ ``` python data = pd.read_csv("https://raw.githubusercontent.com/holtzy/data_to_viz/master/Example_dataset/3_TwoNumOrdered.csv") data[['date', 'value']] = \ data['date value'].str.split(' ', expand=True) # read_csv doesn't always do its job well data.drop(columns=['date value'], inplace=True) # otherwise was string data['value'] = data['value'].astype(float) data['date'] = pd.to_datetime(data['date']) ``` ``` python plt.figure() *plt.plot(data['date'], data['value'], color="#69b3a2", linewidth=2) *plt.axhline(y=5000, color="orange", linewidth=1) plt.annotate("Bitcoin price reached $20k \nat the end of 2017", * xy=(pd.to_datetime('2017-01-01'), 20089), ha='center', va='bottom') plt.ylim(0, 23000) ``` ] .pull-right[ ``` ## (0.0, 23000.0) ``` <img src="14_MakingPlots_files/figure-html/unnamed-chunk-6-1.png" width="672" /> ] --- # Scatter plots (R) - Displays the relation between **two continuous variables** - Works well for reasonably sized data sets - or use **transparency!** .pull-left[ ``` r library(cowplot) ggplot(iris, aes(Sepal.Width, Sepal.Length, color = Species)) + * geom_point(size = 3) + theme_cowplot(16) + theme(legend.position = "bottom") ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-8-1.png" width="90%" /> ] --- # Scatter plots (python) .pull-left[ ``` python import seaborn as sns from matplotlib import pyplot as plt iris = sns.load_dataset("iris") *plt.figure(figsize=(8, 6)) *sns.scatterplot(data=iris, x='sepal_width', y='sepal_length', hue='species', * s=100) # or plt.scatter() # but with manual 'color' and 'label' arguments plt.legend(loc='upper left') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-10-1.png" width="768" /> ] --- # Scatter plots with transparency - With a lot of points, transparency can help to see the density of points .pull-left[ ``` python x = np.random.normal(0, 1, 3000) y = np.random.normal(0, 1, 3000) # no need of seaborn in this simple case plt.scatter(x, y, alpha=0.3, edgecolors='none') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-12-3.png" width="672" /> ] --- # Ellipses can be added to highlight groups - `stat_ellipse` can add confidence ellipses to aid group identities .pull-left[ ``` r library(cowplot) ggplot(iris, aes(Sepal.Width, Sepal.Length, color = Species, fill = Species)) + * stat_ellipse(linewidth = 1) + geom_point(size = 3, shape = 21, color = "black") + theme_cowplot(16) + theme(legend.position = "bottom") ``` - no built-in equivalent in matplotlib/seaborn ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-14-1.png" width="90%" /> ] --- # Trend lines with `geom_smooth` (R) - By default, geom_smooth uses a **loess** curve, a very simple mean trend estimator - [**loess**](https://en.wikipedia.org/wiki/Local_regression) = locally estimated scatterplot smoothing .pull-left[ ``` r ggplot(iris, aes(Sepal.Width, Sepal.Length, color = Species, fill = Species)) + geom_point(size = 3, shape = 21, color = "black") + * geom_smooth(aes(group = Species)) + theme_cowplot(16) + theme(legend.position = "bottom") ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-16-1.png" width="90%" /> ] --- # Trend lines with `statsmodels` (python) - Need to install `statsmodels` for this: `pip install statsmodels` in terminal .pull-left[ ``` python iris = sns.load_dataset("iris") plt.figure() *sns.lmplot(data=iris, x='sepal_width', y='sepal_length', hue='species', * lowess=True) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-18-1.png" width="800" /><img src="14_MakingPlots_files/figure-html/unnamed-chunk-18-2.png" width="592" /> ] --- # Trend lines are bad, rather use linear regression - Smooth trends are unscientific and can be misleading - Rather use linear models for a simple guide-for-the-eye - Linear fit by changing the `method` argument in `geom_smooth` .pull-left[ ``` r ggplot(iris, aes(Sepal.Width, Sepal.Length, color = Species, fill = Species)) + geom_point(size = 3, shape = 21, color = "black") + geom_smooth(aes(group = Species), * method = "lm") + theme_cowplot(16) + theme(legend.position = "bottom") ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-20-1.png" width="90%" /> ] --- # Linear regression in seaborn - Just remove the `lowess=True`! .pull-left[ ``` python iris = sns.load_dataset("iris") plt.figure() *sns.lmplot(data=iris, x='sepal_width', y='sepal_length', hue='species') plt.legend() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-22-1.png" width="800" /><img src="14_MakingPlots_files/figure-html/unnamed-chunk-22-2.png" width="592" /> ] --- class: inverse, center, middle # Distributions of continuous variables --- # Distributions - histograms (R) - Observations from continuous distributions are distributed according to **probability distributions** - We can represent this using histograms or density plots .pull-left[ ``` r x = rnorm(1000, mean = 5, sd = 1) data.frame(x) |> ggplot(aes(x)) + * geom_histogram(fill = "black") + geom_vline(xintercept = mean(x), color = 2, linewidth = 2) + theme_minimal_grid() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-24-1.png" width="90%" /> ] --- # Distributions - histograms (python) .pull-left[ ``` python x = np.random.normal(5, 1, 1000) plt.figure() *plt.hist(x, bins=30, color="black") plt.axvline(np.mean(x), color="red", linewidth=2) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-26-1.png" width="672" /> ] --- # Distributions - density (R) - Observations from continuous distributions are distributed according to **probability distributions** - We can represent this using histograms or density plots .pull-left[ ``` r data.frame(x) |> ggplot(aes(x)) + * geom_density(color = "black", fill = "gray70", alpha = 0.3) + geom_vline(xintercept = mean(x), color = 2, linewidth = 2) + theme_minimal_grid() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-28-1.png" width="90%" /> ] --- # Distributions - density (python) .pull-left[ ``` python plt.figure() *sns.kdeplot(x, color="black", fill=True, alpha=0.3) plt.axvline(np.mean(x), color="red", linewidth=2) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-30-1.png" width="672" /> ] --- # Distributions - histogram and density - Measurements from continuous distributions usually have more or less probable values - We can represent this using histograms or density plots .pull-left[ ``` r data.frame(x) |> ggplot(aes(x)) + * geom_histogram(aes(y = after_stat(density)), fill = "lightblue", bins = 30) + geom_density(color = "black", fill = "gray70", alpha = 0.3, linewidth=2) + geom_vline(xintercept = mean(x), color = 2, linewidth = 2) + theme_minimal_grid() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-32-1.png" width="90%" /> ] --- class: inverse, center, middle # 2D distributions of continuous variables --- # From 2D scatter plots... - Scatter plots can break down if there are too many points -- though transparency might help! .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) ggplot(data, aes(x=x, y=y) ) + * geom_point() + theme_cowplot(16) ``` - python: ``` python plt.scatter(data['x'], data['y'], alpha=0.5) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-35-1.png" width="90%" /> ] --- # To 2D histograms... - So good idea to summarize into bins .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) ggplot(data, aes(x=x, y=y) ) + * geom_bin2d(bins = 70) + scale_fill_viridis() + theme_cowplot(16) ``` - python: ``` python *plt.hist2d(data['x'], data['y'], bins=70, cmap='viridis') plt.colorbar() ``` ] .pull-right[ ``` ## Loading required package: viridisLite ``` <img src="14_MakingPlots_files/figure-html/unnamed-chunk-38-1.png" width="90%" /> ] --- # Or 2D hexagonal histograms ... - Hexagonal bins look nicer than squares .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) ggplot(data, aes(x=x, y=y) ) + * geom_hex(bins = 70) + scale_fill_viridis() + theme_cowplot(16) ``` - python: ``` python *plt.hexbin(data['x'], data['y'], bins=70, cmap='viridis') plt.colorbar() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-41-1.png" width="90%" /> ] --- # Until 2D contour plots - Scatter plots can break down if there are too many points .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) ggplot(data, aes(x=x, y=y) ) + * stat_density_2d(aes(fill = after_stat(level)), geom = "polygon") + scale_fill_viridis() + theme_cowplot(16) ``` - python: ``` python sns.kdeplot(data=data, x="x", y="y", levels=15, fill=True, cmap="viridis") plt.colorbar() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-44-1.png" width="90%" /> ] --- # And 2D density plots - Scatter plots can break down if there are too many points .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) ggplot(data, aes(x=x, y=y) ) + * stat_density_2d(aes(fill = after_stat(density)), * geom = "raster", contour = FALSE) + scale_fill_viridis() + scale_x_continuous(expand = c(0, 0)) + scale_y_continuous(expand = c(0, 0)) + theme_cowplot(16) ``` - python: ``` python # without levels for a raster plot *sns.kdeplot(data=data, x="x", y="y", fill=True, cmap="viridis") plt.colorbar() ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-47-1.png" width="90%" /> ] --- # A last way for 2d density in R - `ggpointdensity` - Scatter plots can break down if there are too many points .pull-left[ ``` r # Data a <- data.frame( x=rnorm(20000, 10, 1.9), y=rnorm(20000, 10, 1.2) ) b <- data.frame( x=rnorm(20000, 14.5, 1.9), y=rnorm(20000, 14.5, 1.9) ) c <- data.frame( x=rnorm(20000, 9.5, 1.9), y=rnorm(20000, 15.5, 1.9) ) data <- rbind(a,b,c) # install.packages("ggpointdensity") library("ggpointdensity") ggplot(data, aes(x=x, y=y) ) + * geom_pointdensity() + scale_color_viridis() + theme_cowplot(16) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-49-1.png" width="90%" /> ] --- class: inverse, center, middle # Continuous distributions vs categorical variables --- # Ridge plots with `ggridges` (R) - Separates distributions according to a categorical variable (here by months) .pull-left[ ``` r library(ggridges); library(viridis) library(hrbrthemes) ggplot(lincoln_weather, aes(x = `Mean Temperature [F]`, y = `Month`, fill = after_stat(x))) + * geom_density_ridges_gradient(scale = 3, rel_min_height = 0.01) + scale_fill_viridis(name = "Temp. [F]", option = "C") + labs(title = 'Temperatures in Lincoln NE') + theme_ipsum() + theme( legend.position="none", panel.spacing = unit(0.1, "lines"), strip.text.x = element_text(size = 8) ) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-51-1.png" width="90%" /> ] --- # Violin plots (Python) - No equivalent of ridge plots in matplotlib or seaborn, but see package `ridgeplot` if needed - **Violin plots** are a "flat" version of ridge plots: a way to visualize multiple distributions .pull-left[ ``` python lincoln_weather = pd.read_csv("https://raw.githubusercontent.com/vega/vega/refs/heads/main/docs/data/seattle-weather.csv") lincoln_weather['month'] = pd.to_datetime( lincoln_weather['date']).dt.month plt.figure() sns.violinplot(data=lincoln_weather, x='month', y='temp_max', scale='width', inner='stick') plt.title('Temperatures in Lincoln NE') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-53-1.png" width="672" /> ] --- # percentiles vs gaussian distributions (boxplots) 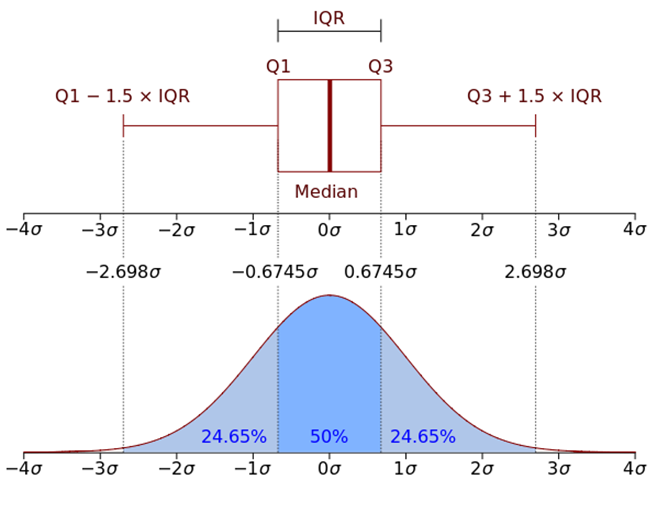 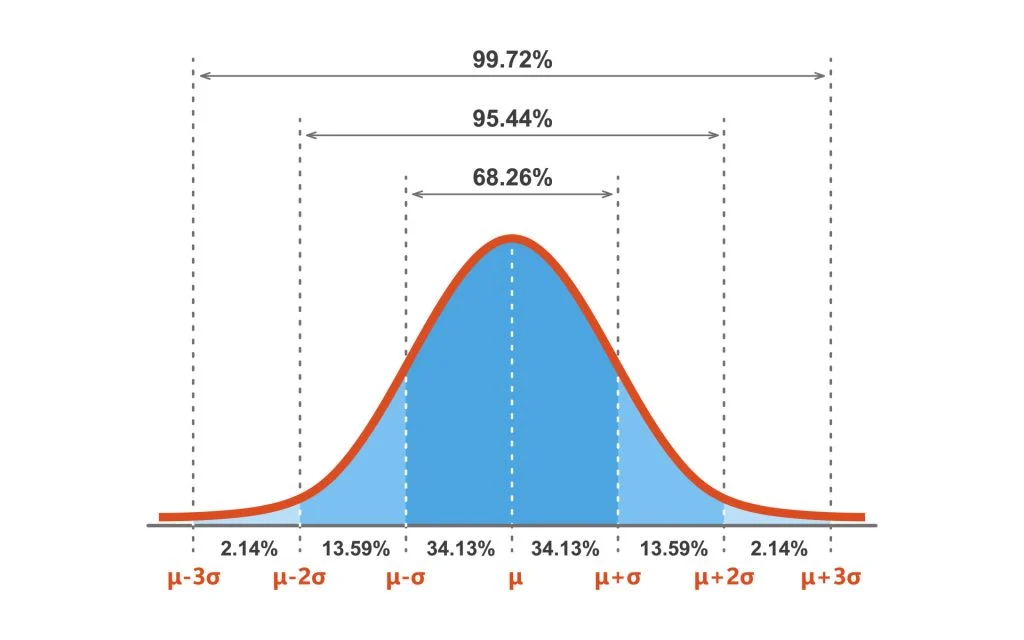 - Interquartile range (IQR) is the `\([25\%, 75\%]\)` box, containing the central `\(50\%\)` of the data - Whiskers are 1.5 times the IQR, containing `\(99.3\%\)` of the (gaussian) data - Other ways to count the confidence intervals: `\(1\sigma = 68.3\%\)` , `\(2\sigma = 95.4\%\)`, `\(3\sigma = 99.7\%\)` --- # Boxplots - Relates continuous and categorical variables, as violin and ridge plots, but with more **synthetic information on distributions** - Allows for comparisons of main **quantiles** across groups .pull-left[ ``` r ggplot(ToothGrowth, aes(x=factor(dose), y=len)) + * geom_boxplot(fill = 2) + labs(x = "Dose", y = "Length") + theme_cowplot(16) ``` - seaborn equivalent: ``` python *sns.boxplot(data=ToothGrowth, x='dose', y='len') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-56-1.png" width="90%" /> ] --- # Jitter plots (without jitter) - Equivalent of **scatter plots for continuous vs categorical** - To use when **exhaustivity on all data points** is important .pull-left[ ``` r ggplot(ToothGrowth, aes(x=factor(dose), y=len)) + geom_point() + labs(x = "Dose", y = "Length") + theme_cowplot(16) ``` - python: simply ``` python sns.scatterplot(data=ToothGrowth, x='dose', y='len') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-59-1.png" width="90%" /> ] --- # Jitter plots - Equivalent of **scatter plots for continuous vs categorical** - To use when **exhaustivity on all data points** is important - Can add some **jitter** to avoid overlap .pull-left[ ``` r ggplot(ToothGrowth, aes(x=factor(dose), y=len)) + * geom_jitter(width = 0.1, height = 0) + labs(x = "Dose", y = "Length") + theme_cowplot(16) ``` - python: in seaborn ``` python *sns.stripplot(data=ToothGrowth, x='dose', y='len', jitter=True) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-62-1.png" width="90%" /> ] # Jitter plots and boxplots .pull-left[ ``` r ggplot(ToothGrowth, aes(x=factor(dose), y=len)) + * geom_boxplot(fill = 2, outlier.alpha = 0) + * geom_jitter(width = 0.1, height = 0, size = 2) + labs(x = "Dose", y = "Length") + theme_cowplot(16) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-64-1.png" width="90%" /> ] --- # Barplots with uncertainty bars (R) - Relates mean measurements with a categorical variable - Some argue it should be used when zero is a value of interest... - But sometimes **excluding the value zero is misleading the reader in terms of magnitude of the effect!** .pull-left[ ``` r iris |> select(Species, Sepal.Length) |> group_by(Species) |> summarise(n=n(), mean=mean(Sepal.Length), * sd=sd(Sepal.Length) ) |> ggplot( aes(x=Species, y=mean)) + * geom_bar(stat="identity", fill="forestgreen", alpha=0.5) + * geom_errorbar(aes(ymin=mean-sd, ymax=mean+sd), width=0.4, colour="gray30", alpha=0.9, linewidth=1.5) + scale_y_continuous (expand = expansion(mult = c(0, 0.05))) + labs(caption = "Using standard deviation") + theme_cowplot(16) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-66-1.png" width="90%" /> ] --- # Barplots with uncertainty bars (python) - Relates mean measurements with a categorical variable - Some argue it should be used when zero is a value of interest... - But sometimes **excluding the value zero is misleading the reader in terms of magnitude of the effect!** .pull-left[ ``` python iris = sns.load_dataset("iris") summary = iris.groupby('species').agg( mean=('sepal_length', 'mean'), * se=('sepal_length', 'std') ).reset_index() plt.bar(summary['species'], summary['mean'], color='forestgreen', alpha=0.5) *plt.errorbar(summary['species'], summary['mean'], * yerr=summary['se'], fmt='o', color='gray', capsize=5) plt.xlabel("Species") plt.ylabel("Mean Sepal Length") ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-68-1.png" width="90%" /> ] --- # Point range plots - Great alternative to barplots when zero is not special .pull-left[ ``` r iris |> select(Species, Sepal.Length) |> group_by(Species) |> summarise(n=n(), mean=mean(Sepal.Length), sd=sd(Sepal.Length) ) |> ggplot( aes(x=Species, y=mean)) + geom_point() + geom_errorbar(aes(ymin=mean-sd, ymax=mean+sd), width=0.1, colour="gray30", alpha=0.9, linewidth=1.5) + labs(title = "Using standard deviation") + theme_cowplot(16) ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-70-1.png" width="90%" /> ] --- # Barplots vs point range .pull-left[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-71-1.png" width="90%" /> ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-72-1.png" width="90%" /> ] --- class: inverse, center, middle # Categorical vs categorical --- # Cleveland plots (R) - Good alternative to barplots if you want to contrast two groups in many different conditions .pull-left[ ``` r smoking = read_csv("https://raw.githubusercontent.com/EEB330/slides/data/oecd_smoking.csv") smoking |> mutate(Country = factor(Country, # This maintains the row order in the plot unique(Country))) |> pivot_longer(Men:Women, names_to = "Gender", values_to = "proportion") |> ggplot(aes(Country, proportion, color = Gender)) + geom_point(size = 5) + geom_line(aes(group = Country)) + coord_flip() + scale_color_manual(values = 1:2) + labs(x = NULL, y = "Proportion of smokers", caption = "https://www.oecd-ilibrary.org/") + theme_minimal_grid(16) + theme(legend.position = c(0.8, 0.1), legend.box.background = element_rect(color = "white", fill = "white")) ``` ] .pull-right[ ``` ## Warning: A numeric `legend.position` argument in `theme()` was deprecated in ggplot2 3.5.0. ## ℹ Please use the `legend.position.inside` argument of `theme()` instead. ## This warning is displayed once every 8 hours. ## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated. ``` <img src="14_MakingPlots_files/figure-html/unnamed-chunk-74-1.png" width="90%" /> ] --- # Cleveland plots (python) - Good alternative to barplots if you want to contrast two groups in many different conditions .pull-left[ ``` python smoking = pd.read_csv("data/oecd_smoking.csv") smoking_long = smoking.melt( id_vars="Country", value_vars=["Men", "Women"], var_name="Gender", value_name="proportion") plt.figure() # Plot points for each gender with separate colors sns.scatterplot(data=smoking_long, x="proportion", y="Country", hue='Gender') # Draw lines connecting the points for each country for country in smoking_long["Country"].unique(): subset = smoking_long[smoking_long["Country"] == country] plt.plot(subset["proportion"], subset["Country"], color='gray', linestyle='-', linewidth=1) plt.title("Proportion of Smokers by Country and Gender") plt.xlabel("Proportion of smokers") plt.ylabel("") plt.legend(title="Gender") plt.grid(True, which='both', axis='x') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-76-1.png" width="672" /> ] --- # Heatmaps (categorical) - Can be used e.g. for representing correlation matrices - Also common for showing all points across some condition (RNAseq, microarray...) - R package: [`ggheatmap`](https://github.com/XiaoLuo-boy/ggheatmap) .pull-left[ ``` r cor_matrix = cor(iris[,-5]) diag(cor_matrix) = 0 melted_cormat = reshape2::melt(cor_matrix) ggplot(melted_cormat, aes(x=Var1, y=Var2, fill=value)) + geom_tile() + labs(x = NULL, y = NULL, fill = "Correlation") + scale_fill_distiller(type="div", palette="RdBu", limits = c(-1, 1)) + ``` - python: ``` python corr_matrix = iris.iloc[:, :-1].corr() *sns.heatmap(corr_matrix, annot=True, cmap='RdBu') ``` ] .pull-right[ <img src="14_MakingPlots_files/figure-html/unnamed-chunk-79-1.png" width="85%" /> ] --- class: left, top background-image: url(figures/ggplot_ext.png) background-position: center right background-size: 100% # [ggplot2 extension gallery](https://exts.ggplot2.tidyverse.org/gallery/)